
First, in the global mode, the alignments in the whole genome are drawn in one or more continuous pages to show the users an overall situation ( Fig.
#Laboratory song software#
RAviz is a Windows- or MacOS-based open-source software that is able to visualize the alignments between any types of DNA sequences such as reads, contigs, scaffolds or reference genomes in three different modes ( Fig. With the alignments and corresponding k-mer matching profiles clearly visualized by RAviz, it is much easier and time-efficient for the users to decide which are the false alignments and remove them in T2T assembly projects.
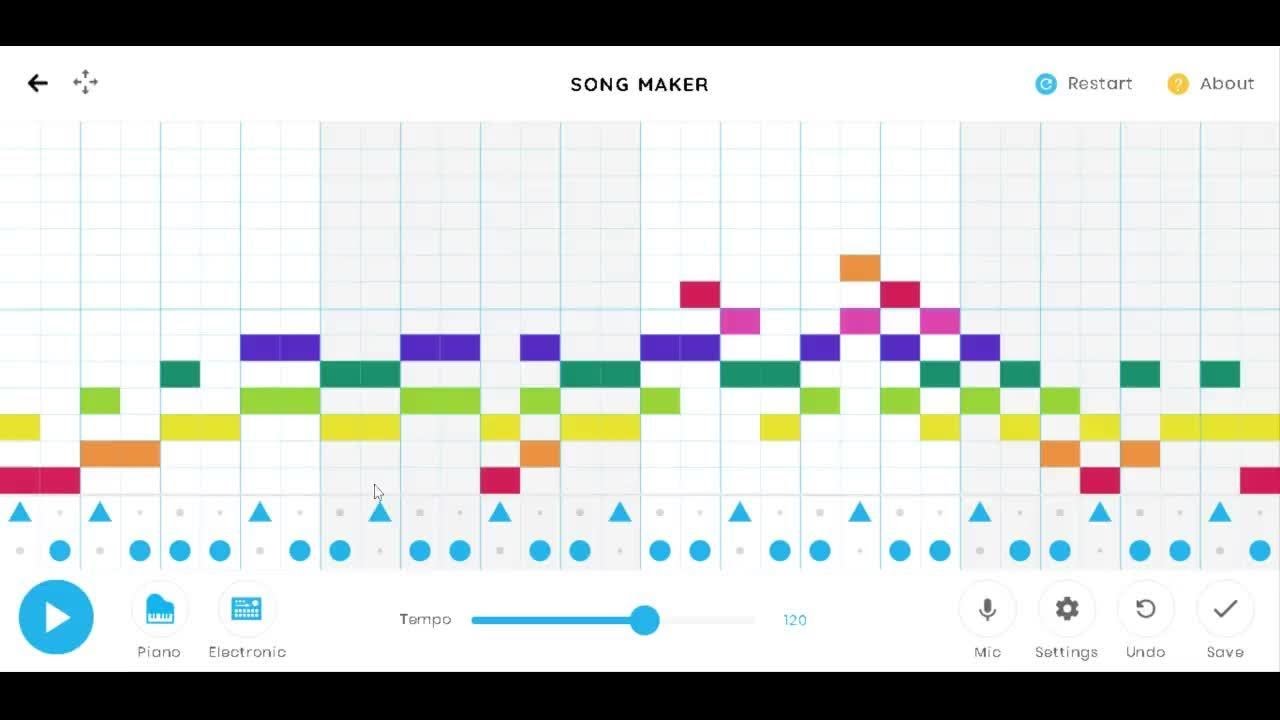
Here we have developed an efficient alignment visualization tool called RAviz to meet this need. This is the reason that other T2T or near-T2T projects choose not to or use a rougher strategy to filter false-positive alignments, leading to lower correctness than human T2T assembly. However, due to the lack of a visualization tool that can show the match of rare k-mers in alignments (the existing alignment visualization tools such as IGV show the alignments without k-mer matching), it is extremely tedious and time consuming to carry out this strategy manually.
#Laboratory song manual#
This strategy has been widely used by T2T-related automatic tools like CentroFlye, Abruijn, and manual human T2T assembly. Due to the difficulty and high computational cost of SNP and Structural Variation (SV) detection, in practice, people use rare k-mers (subsequences of k nucleotides appearing in the whole genome for only a small number of times) as markers to replace these copy-specific features, and the alignments with rare k-mers mismatching should be removed. More specifically, the alignments in which the copy-specific features from two sequences do not match should be identified as false-positive. To filter these false-positive alignments and ensure the correctness of repeat assembly, copy-specific features like Single Nucleotide Polymorphisms (SNPs) and structural variations have to be used. However, due to the high similarity between the repeat copies, the sequences from different copies may be aligned by mistake, which leads to mis-assemblies in these steps. These manual works focus on generating continuous and correct sequences in repetitive regions in some steps of genome assembly such as contig assembly, scaffolding, polishing and gap filling. Fortunately, the generation of Pacbio high-fidelity (HiFi) long sequences and Oxford Nanopore Technology (ONT) ultra-long (UL) sequences provides the opportunity of solving repeat assembly problems because of their advantages in accuracy and length, respectively, and a list of complete (T2T, or Telomere to Telomere) or near-complete reference genomes of important eukaryotic species like human, Arabidopsis, rice and tomato have been built recently.īecause there has been no assembler that can generate complete reference genomes purely automatically until now, these T2T projects all need large amounts of manual curations. The incomplete reference genomes not only cause data analytical mistakes like false-positive variant calls but also impede the studies of repeat-related diseases such as cancer and infertility.
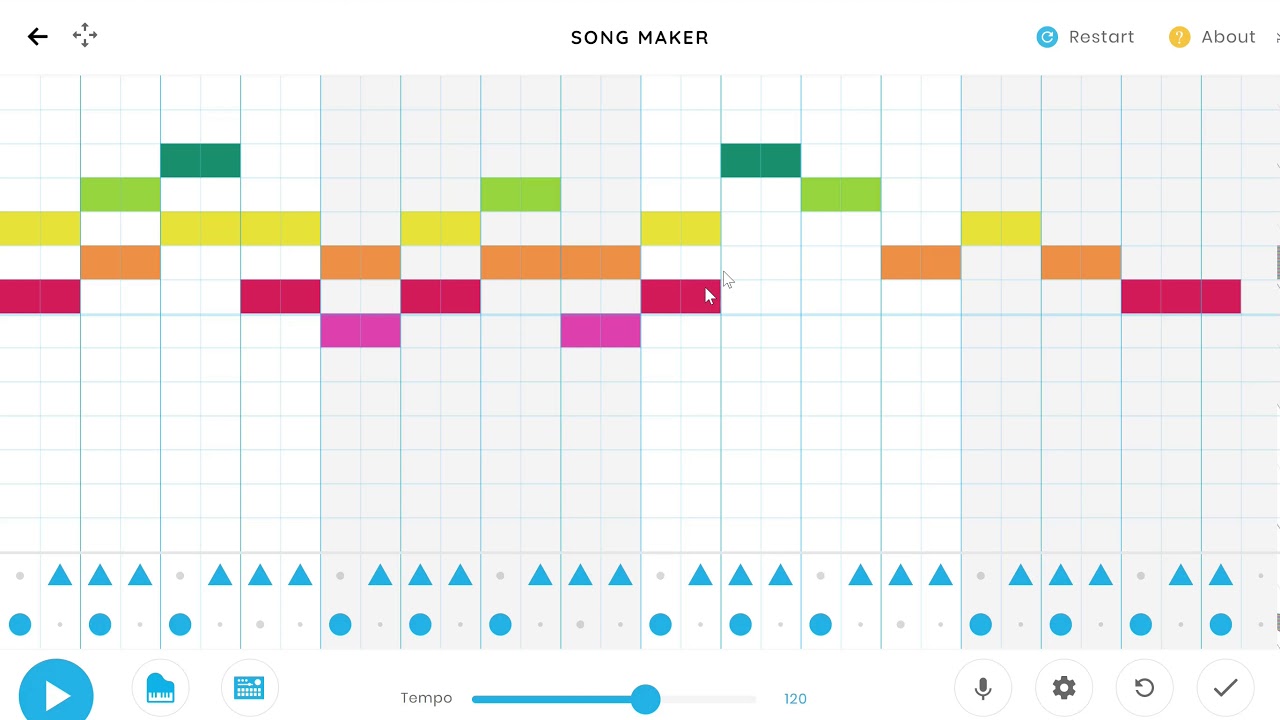

However, for decades the reference sequences of important eukaryotic genomes were incomplete due to the missing repetitive genomic regions including both tandem repeats such as centromere, telomere, and ribosomal DNA, and interspersed repeats like transposons and segmental duplications. For any species, a high-quality reference genome is the basis for almost all kinds of genomic analysis.


 0 kommentar(er)
0 kommentar(er)
